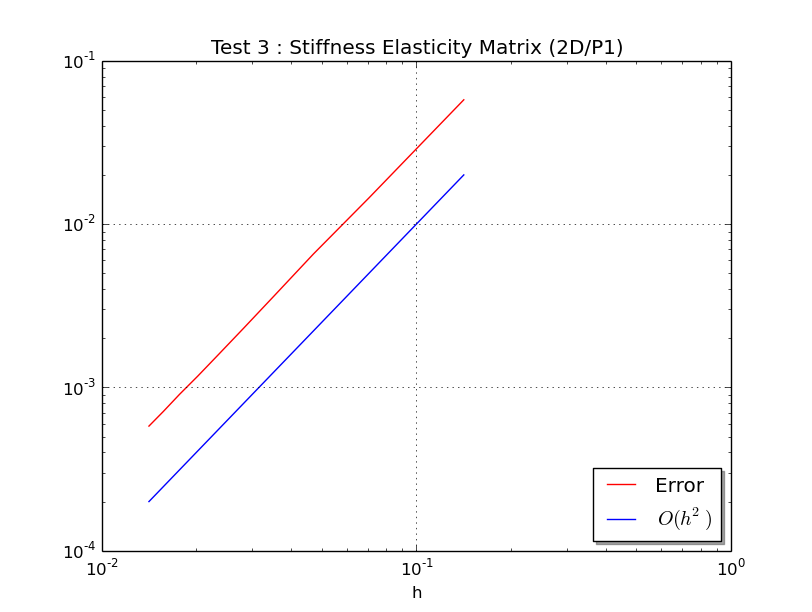
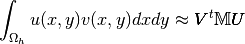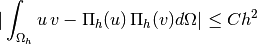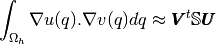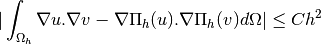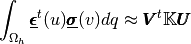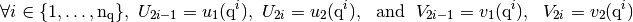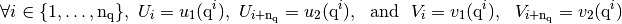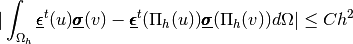valid2D module¶
Mass Matrix¶
Validation program for the assembly of the Mass matrix 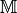 for
for 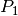 -Lagrange finite element method in 2d
(see Mass Matrix).
-Lagrange finite element method in 2d
(see Mass Matrix).
Test 1: Computation of the Mass Matrix using the base, OptV1 and OptV2 versions : it gives errors and computation times
Test 2: Computation of the integral
where
and
. Functions
and
are those defined in ...
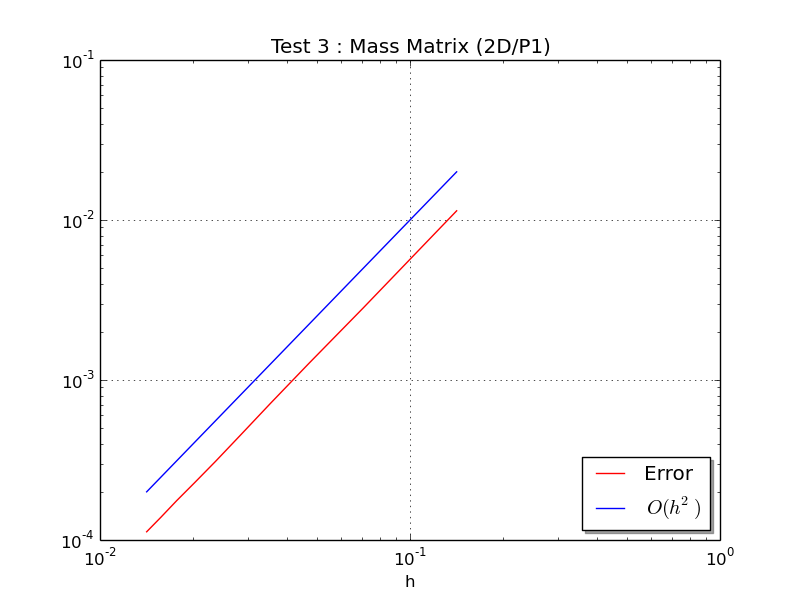
Test 3: Ones retrieves the order 2 of
-Lagrange integration
Note
source code
>>> from pyOptFEM.valid2D import validMass2DP1
>>> validMass2DP1()
******************************************
* Mass Assembling P1 validations *
******************************************
-----------------------------------------
Test 1: Matrices errors and CPU times
-----------------------------------------
Matrix size : (121,121)
Error P1base vs OptV1 : 8.673617e-19
Error P1base vs OptV2 : 8.673617e-19
CPU times base (ref) : 0.2638 (s)
CPU times OptV1 : 0.0259 (s) - Speed Up X10.179
CPU times OptV2 : 0.0016 (s) - Speed Up X160.286
----------------------------
Test 1 (results): OK
----------------------------
-----------------------------------------------------
Test 2: Validations by integration on [0,1]x[0,1]
-----------------------------------------------------
function 0 : u(x,y)=x+2*y, v(x,y)=3*x+y+1,
-> Mass error=0.000000e+00
function 1 : u(x,y)=x**2+2*y*x+y, v(x,y)=3*x*y+y**2+1,
-> Mass error=5.736389e-03
function 2 : u(x,y)=x**3+2*y**2*x+y**2+x, v(x,y)=2*x*y+y**3+x*y,
-> Mass error=1.141472e-02
----------------------------
Test 2 (results): OK
----------------------------
--------------------------------
Test 3: Validations by order
--------------------------------
functions 2: u(x,y)=x**3+2*y**2*x+y**2+x, v(x,y)=2*x*y+y**3+x*y
Matrix size : (121,121)
MassAssemblingP1OptV2 CPU times : 0.001(s)
Error : 1.141472e-02
Matrix size : (441,441)
MassAssemblingP1OptV2 CPU times : 0.002(s)
Error : 2.825605e-03
...
Matrix size : (10201,10201)
MassAssemblingP1OptV2 CPU times : 0.019(s)
Error : 1.125457e-04
At last, this program plots the figure :
Stiffness matrix¶
Validation function for the assembly of the Stiffness matrix, 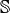 , for
, for 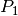 -Lagrange
finite element method in 2d (see Stiffness Matrix)
-Lagrange
finite element method in 2d (see Stiffness Matrix)
Test 1: Computation of the Stiffness Matrix using the base, OptV1 and OptV2 versions : it gives errors and computation times
Test 2: Computation of the integral
where
and
. Functions
and
are those defined in ...
Test 3: Ones retrieves the order 2 of
-Lagrange integration
Note
source code
- pyOptFEM.valid2D.validStiff2DP1.validStiff2DP1(**kwargs)[source]
validation of stiffness matrix assembling functions
>>> from pyOptFEM.valid2D import validStiff2DP1
>>> validStiff2DP1()
**********************************************
* 2D Stiff Assembling P1 validations *
**********************************************
-----------------------------------------
Test 1: Matrices errors and CPU times
-----------------------------------------
Matrix size : (121,121)
Error P1base vs OptV1 : 8.881784e-16
Error P1base vs OptV2 : 4.440892e-16
CPU times base (ref) : 0.1519 (s)
CPU times OptV1 : 0.0106 (s) - Speed Up X14.270
CPU times OptV2 : 0.0009 (s) - Speed Up X174.238
----------------------------
Test 1 (results): OK
----------------------------
-----------------------------------------------------
Test 2: Validations by integration on [0,1]x[0,1]
-----------------------------------------------------
function 0 : u(x,y)=x+2*y, v(x,y)=3*x+y+1,
-> Stiff error=2.486900e-14
function 1 : u(x,y)=x**2+2*y*x+y, v(x,y)=3*x*y+y**2+1,
-> Stiff error=2.000000e-02
function 2 : u(x,y)=x**3+2*y**2*x+y**2+x, v(x,y)=2*x*y+y**3+x*y,
-> Stiff error=1.500000e-02
----------------------------
Test 2 (results): OK
----------------------------
--------------------------------
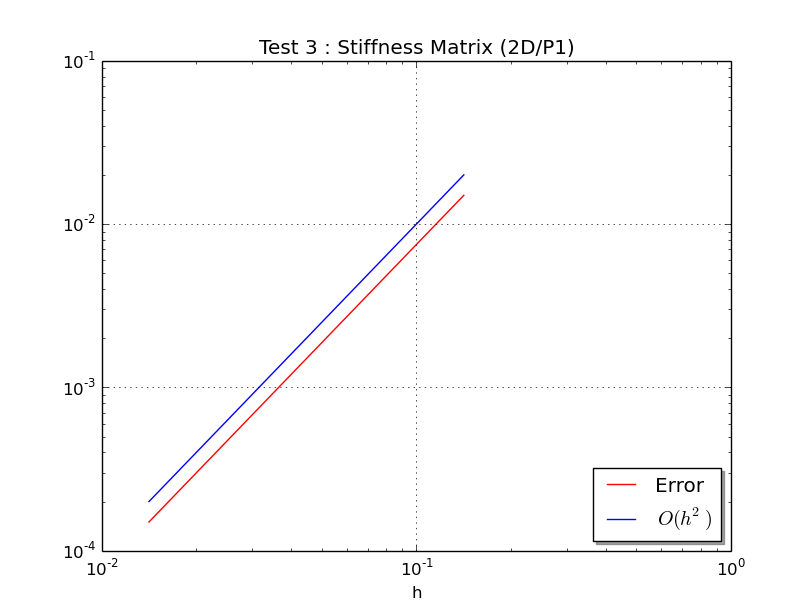
Test 3: Validations by order
--------------------------------
Test 2: u(x,y)=x**3+2*y**2*x+y**2+x, v(x,y)=2*x*y+y**3+x*y
Matrix size : (121,121)
StiffAssemblingP1OptV2 CPU times : 0.001(s)
Error : 1.500000e-02
...
Matrix size : (10201,10201)
StiffAssemblingP1OptV2 CPU times : 0.021(s)
Error : 1.500000e-04
At last, this program plots this figure :
Elastic Stiffness Matrix¶
Validation function for the assembly of the Elastic Stiffness matrix, 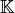 , for
, for 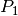 -Lagrange
finite element method in 2d (see Elastic Stiffness Matrix)
-Lagrange
finite element method in 2d (see Elastic Stiffness Matrix)
Test 1: Computation of the Elastic Stiffness Matrix using the base, OptV1 and OptV2 versions : it gives errors and computation times
Test 2: Computation of the integral
where
and
are given 2d-vector functions and
and
are the vectors in
defined by
if Num=0 or Num=2 (i.e. in global alternate basis
, see Assembly matrices (base, OptV1 and OptV2 versions) )
if Num=1 or Num=3 (i.e. in global block basis
, see Assembly matrices (base, OptV1 and OptV2 versions) )
Test 3: Ones retrieves the order 2 of
-Lagrange integration
Note
- pyOptFEM.valid2D.validStiff2DP1.validStiffElas2DP1([Num=0, la=1.5, mu=0.5])¶
Parameters: - Num – (optional) Numbering choice.
- la – (optional) the first Lame coefficient in Hooke’s law, denoted by
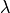 .
. - mu – (optional) the second Lame coefficient in Hooke’s law, denoted by
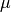 .
.
- pyOptFEM.valid2D.validStiffElas2DP1.validStiffElas2DP1(**kwargs)[source]
validation of elasticity stiffness matrix assembly functions
>>> from pyOptFEM.valid2D import validStiffElas2DP1
>>> validStiffElas2DP1()
**************************************************
* 2D StiffElas Assembling P1 validations *
**************************************************
Numbering Choice : global alternate numbering with local alternate numbering
-----------------------------------------
Test 1: Matrices errors and CPU times
-----------------------------------------
Matrix size : (1922,1922)
Error P1base vs OptV1 : 1.776357e-15
Error P1base vs OptV2 : 2.664535e-15
CPU times base (ref) : 1.9251 (s)
CPU times OptV1 : 0.2817 (s) - Speed Up X6.835
CPU times OptV2 : 0.0098 (s) - Speed Up X195.725
----------------------------
Test 1 (results): OK
----------------------------
-----------------------------------------------------
Test 2: Validations by integration on [0,1]x[0,1]
-----------------------------------------------------
function 0 :
u(x,y)=[x - 2*y,x + y],
v(x,y)=[x + 2*y,2*x - y],
-> StiffElas error=8.704149e-14
function 1 :
u(x,y)=[x**2+2*y*x+y,-2*y**2+x**2+x-y],
v(x,y)=[3*x*y+y**2+1,3*x**2-x*y+1],
-> StiffElas error=3.916049e-03
function 2 :
u(x,y)=[x**3+2*y**2*x+y**2+x,y**3-2*x**2*y],
v(x,y)=[2*x*y+y**3+x*y,3*x**3-2*x*y+x-1],
-> StiffElas error=6.574856e-03
----------------------------
Test 2 (results): OK
----------------------------
--------------------------------
Test 3: Validations by order
--------------------------------
functions 2:
u(x,y)=['x**3+2*y**2*x+y**2+x', 'y**3-2*x**2*y'],
v(x,y)=['2*x*y+y**3+x*y', '3*x**3-2*x*y+x-1'],
lambda=1.500000, mu=0.500000
Matrix size : (242,242)
StiffElasAssemblingP1OptV2 CPU times : 0.002(s)
Error : 5.767667e-02
Matrix size : (882,882)
StiffElasAssemblingP1OptV2 CPU times : 0.005(s)
Error : 1.447542e-02
Matrix size : (1922,1922)
...
Matrix size : (20402,20402)
StiffElasAssemblingP1OptV2 CPU times : 0.110(s)
Error : 5.797263e-04
At last, this program plots this figure :