2D benchmarks¶
Benchmark usage¶
- pyOptFEM.FEM2D.assemblyBench.assemblyBench([assembly=<string>, version=<string>, LN=<array>, meshname=<string>, meshdir=<string>, nbruns=<int>, la=<float>, mu=<float>, Num=<int>, tag=<string>, ...])
Benchmark code for
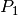 -Lagrange finite elements Matrices defined in FEM2D.
-Lagrange finite elements Matrices defined in FEM2D.Parameters: - assembly – Name of an assembly routine. String must be : - ‘MassAssembling2DP1’, - ‘StiffAssembling2DP1’ (default) - ‘StiffElasAssembling2DP1’
- versions – Versions list. Must be an array of in {‘base’,’OptV1’,’OptV2’} (default : ['OptV2','OptV1','base'])
- meshname – Name of the mesh files. By default is empty string ‘’ : use SquareMesh(N) function to generate meshes.
- meshdir – Directory location of FreeFEM++ mesh files. Used if meshname is not empty.
- LN –
array of integer values. Used to generate meshes data via Th=SquareMesh(N) function if meshname is empty or via getMesh class to read FreeFEM++ meshes Th=getMesh(meshdir+’/’+meshname+str(N)+’.msh’)
Default is range(50, 90, 10)
- nbruns – Number of run on each mesh (default : 1)
- save – Save benchmark results in a file (see params <output>, <outdir> and <tag>)
- plot – (default : True)
- output – Name used to set results file name (default : ‘bench2D’)
- outdir – Directory location of saved results file name (default : ‘./results’).
- tag – Specify a tag string added to filename
- la – the first Lame coefficient in Hooke’s law, denoted by
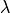 . Used by stiffness elasticity assembly matrix functions
(default : 2)
. Used by stiffness elasticity assembly matrix functions
(default : 2) - mu – the second Lame coefficient in Hooke’s law, denoted by
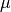 . Used by stiffness elasticity assembly matrix functions.
(default : 0.3)
. Used by stiffness elasticity assembly matrix functions.
(default : 0.3) - Num – Numbering choice. Used by stiffness elasticity assembly matrix functions. (default : 0)
Mass Matrix¶
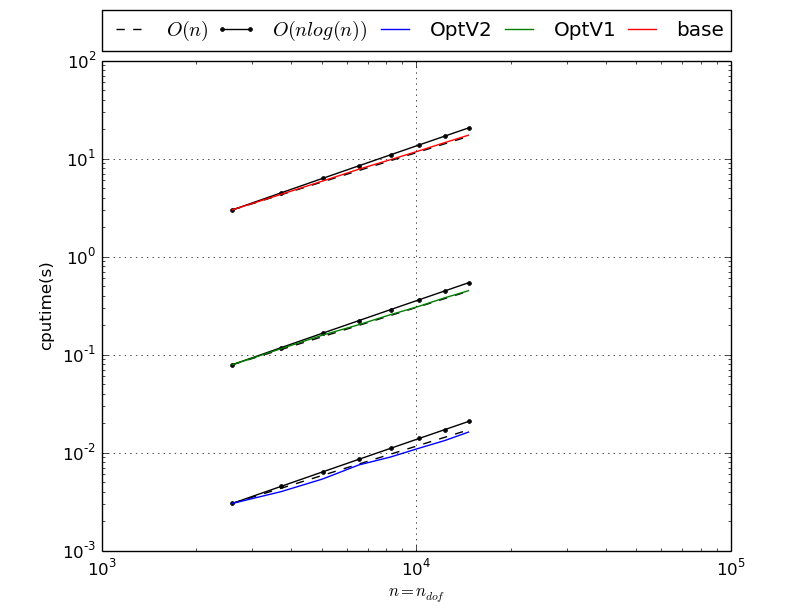
Benchmark of MassAssembling2DP1 with versions base, OptV1 and OptV2 (see Mass Matrix)
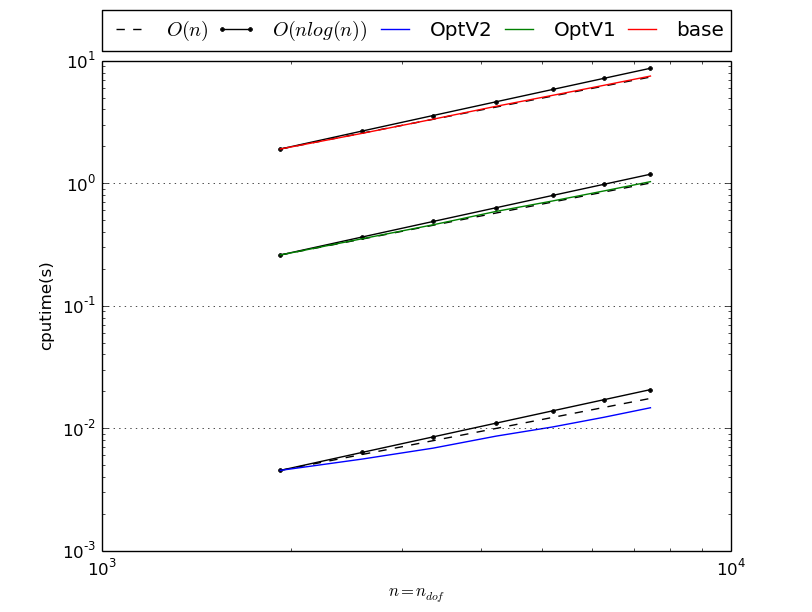
MassAssembling2DP1 benchmark resume N nq ndof OptV2 OptV1 base 50 2601 2601 0.0042(s) 0.0816(s) 3.2783(s) x1.00 x19.50 x783.85 60 3721 3721 0.0058(s) 0.1162(s) 4.7136(s) x1.00 x20.02 x812.43 70 5041 5041 0.0078(s) 0.1584(s) 6.3931(s) x1.00 x20.32 x819.97 80 6561 6561 0.0102(s) 0.2075(s) 8.4154(s) x1.00 x20.33 x824.26 90 8281 8281 0.0133(s) 0.2622(s) 10.6460(s) x1.00 x19.66 x798.28 100 10201 10201 0.0162(s) 0.3228(s) 13.1028(s) x1.00 x19.89 x807.38 110 12321 12321 0.0197(s) 0.3912(s) 15.9244(s) x1.00 x19.88 x809.23 120 14641 14641 0.0232(s) 0.4685(s) 18.9165(s) x1.00 x20.21 x816.09 This tabular was build with the following code :
>>> from pyOptFEM.FEM2D import assemblyBench >>> assemblyBench(assembly='MassAssembling2DP1',LN=range(50,130,10)) SquareMesh(50): -> MassAssembling2DP1OptV2(nq=2601, nme=5000) run ( 1/ 1) : cputime=0.004182(s) - matrix 2601-by-2601 ... SquareMesh(120): -> MassAssembling2DP1base(nq=14641, nme=28800) run ( 1/ 1) : cputime=18.916504(s) - matrix 14641-by-14641 ...
We also obtain
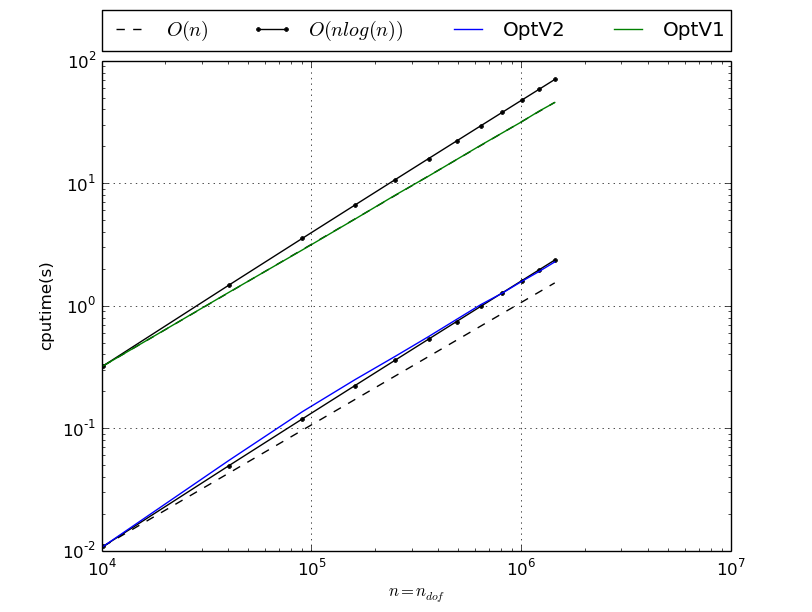
Benchmark of MassAssembling2DP1 with versions OptV2 and OptV1
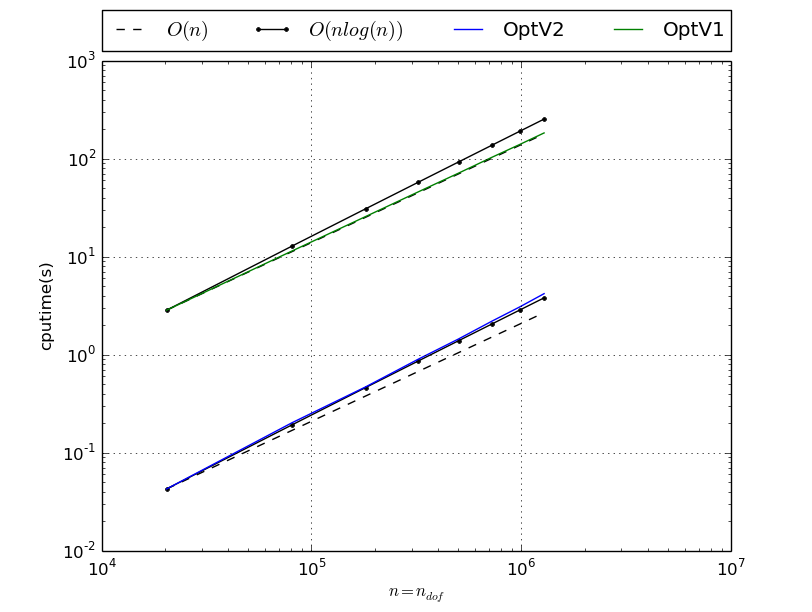
MassAssembling2DP1 benchmark resume N nq ndof OptV2 OptV1 100 10201 10201 0.0163(s) 0.3196(s) x1.00 x19.61 200 40401 40401 0.0648(s) 1.3025(s) x1.00 x20.09 300 90601 90601 0.1583(s) 2.9198(s) x1.00 x18.45 400 160801 160801 0.2859(s) 5.1809(s) x1.00 x18.12 500 251001 251001 0.4591(s) 8.1059(s) x1.00 x17.65 600 361201 361201 0.6595(s) 11.6834(s) x1.00 x17.72 700 491401 491401 0.9056(s) 15.8209(s) x1.00 x17.47 800 641601 641601 1.1796(s) 20.8883(s) x1.00 x17.71 900 811801 811801 1.4933(s) 26.0264(s) x1.00 x17.43 1000 1002001 1002001 1.8503(s) 32.5076(s) x1.00 x17.57 1100 1212201 1212201 2.2262(s) 39.3237(s) x1.00 x17.66 1200 1442401 1442401 2.6616(s) 46.6362(s) x1.00 x17.52 This tabular was build with the following code :
>>> from pyOptFEM.FEM2D import assemblyBench >>> assemblyBench(assembly='MassAssembling2DP1',versions=['OptV2','OptV1'],LN=range(100,1300,100)) SquareMesh(100): -> MassAssembling2DP1OptV2(nq=10201, nme=20000) run ( 1/ 1) : cputime=0.016294(s) - matrix 10201-by-10201 ... SquareMesh(1200): -> MassAssembling2DP1OptV1(nq=1442401, nme=2880000) run ( 1/ 1) : cputime=46.636235(s) - matrix 1442401-by-1442401 ...
We also obtain
Stiffness Matrix¶
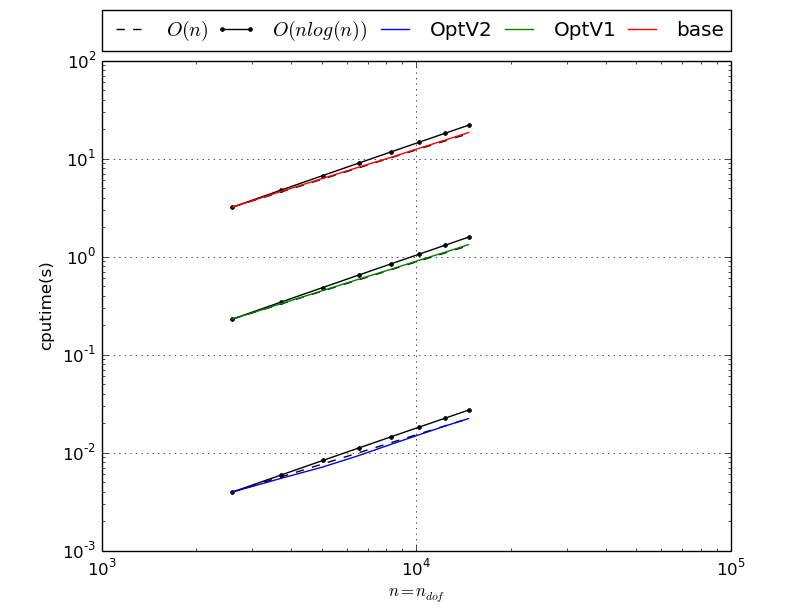
Benchmark of StiffAssembling2DP1 with versions base, OptV1 and OptV2
StiffAssembling2DP1 benchmark resume N nq ndof OptV2 OptV1 base 50 2601 2601 0.0055(s) 0.2501(s) 3.5504(s) x1.00 x45.35 x643.82 60 3721 3721 0.0076(s) 0.3593(s) 5.1301(s) x1.00 x47.38 x676.64 70 5041 5041 0.0101(s) 0.4897(s) 6.9854(s) x1.00 x48.33 x689.37 80 6561 6561 0.0138(s) 0.6362(s) 9.0807(s) x1.00 x46.11 x658.09 90 8281 8281 0.0178(s) 0.8119(s) 11.5354(s) x1.00 x45.57 x647.46 100 10201 10201 0.0223(s) 1.0005(s) 14.2190(s) x1.00 x44.77 x636.22 110 12321 12321 0.0266(s) 1.2124(s) 17.2704(s) x1.00 x45.61 x649.74 120 14641 14641 0.0325(s) 1.4503(s) 20.4368(s) x1.00 x44.69 x629.75 This tabular was build with the following code :
>>> from pyOptFEM.FEM2D import assemblyBench >>> assemblyBench(assembly='StiffAssembling2DP1',LN=range(50,130,10)) SquareMesh(50): -> StiffAssembling2DP1OptV2(nq=2601, nme=5000) run ( 1/ 1) : cputime=0.005515(s) - matrix 2601-by-2601 ... SquareMesh(120): -> StiffAssembling2DP1base(nq=14641, nme=28800) run ( 1/ 1) : cputime=20.436779(s) - matrix 14641-by-14641 ...
We also obtain
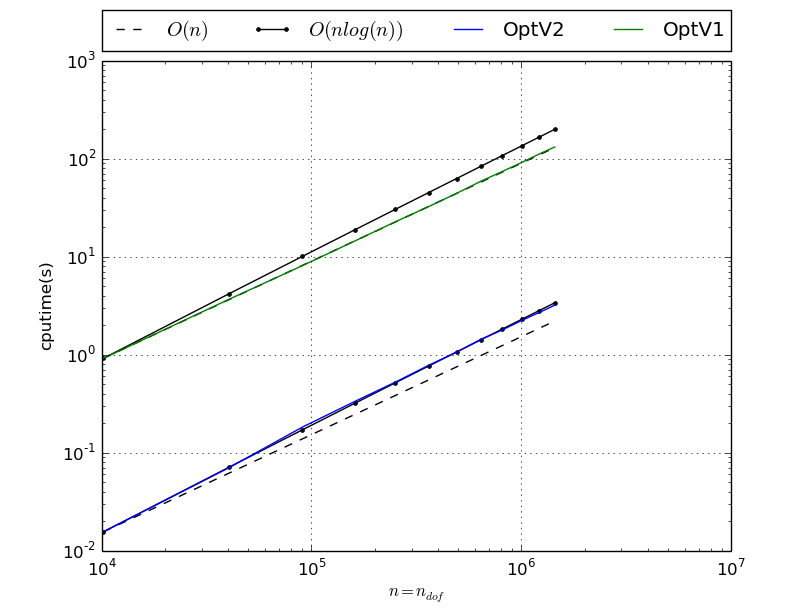
Benchmark of StiffAssembling2DP1 with versions OptV2 and OptV1
StiffAssembling2DP1 benchmark resume N nq ndof OptV2 OptV1 100 10201 10201 0.0225(s) 1.0104(s) x1.00 x44.93 200 40401 40401 0.0945(s) 4.0847(s) x1.00 x43.22 300 90601 90601 0.2379(s) 9.1818(s) x1.00 x38.59 400 160801 160801 0.4459(s) 16.3333(s) x1.00 x36.63 500 251001 251001 0.7169(s) 25.6513(s) x1.00 x35.78 600 361201 361201 1.0316(s) 36.8382(s) x1.00 x35.71 700 491401 491401 1.4316(s) 49.9507(s) x1.00 x34.89 800 641601 641601 1.8965(s) 65.8354(s) x1.00 x34.71 900 811801 811801 2.4164(s) 82.7545(s) x1.00 x34.25 1000 1002001 1002001 2.9450(s) 102.3931(s) x1.00 x34.77 1100 1212201 1212201 3.4908(s) 123.9802(s) x1.00 x35.52 1200 1442401 1442401 4.3144(s) 147.2963(s) x1.00 x34.14 This tabular was build with following code :
>>> from pyOptFEM.FEM2D import assemblyBench >>> assemblyBench(assembly='StiffAssembling2DP1',versions=['OptV2','OptV1'],LN=range(100,1300,100)) SquareMesh(100): -> StiffAssembling2DP1OptV2(nq=10201, nme=20000) run ( 1/ 1) : cputime=0.022488(s) - matrix 10201-by-10201 ... SquareMesh(1200): -> StiffAssembling2DP1OptV1(nq=1442401, nme=2880000) run ( 1/ 1) : cputime=147.296321(s) - matrix 1442401-by-1442401 ...
We also obtain
Stiffness Elasticity Matrix¶
Benchmark of StiffElasAssembling2DP1 with versions base, OptV1 and OptV2
StiffElasAssembling2DP1 benchmark resume N nq ndof OptV2 OptV1 base 30 961 1922 0.0107(s) 0.2809(s) 1.8485(s) x1.00 x26.29 x173.04 35 1296 2592 0.0134(s) 0.3807(s) 2.5109(s) x1.00 x28.37 x187.11 40 1681 3362 0.0172(s) 0.4986(s) 3.2952(s) x1.00 x28.92 x191.13 45 2116 4232 0.0216(s) 0.6306(s) 4.1589(s) x1.00 x29.26 x192.97 50 2601 5202 0.0269(s) 0.7800(s) 5.1323(s) x1.00 x29.04 x191.07 55 3136 6272 0.0322(s) 0.9453(s) 6.2180(s) x1.00 x29.39 x193.35 60 3721 7442 0.0386(s) 1.1282(s) 7.4995(s) x1.00 x29.21 x194.14 This tabular was build with following code :
>>> from pyOptFEM.FEM2D import assemblyBench >>> assemblyBench(assembly='StiffElasAssembling2DP1',LN=range(30,65,5)) SquareMesh(30): -> StiffElasAssembling2DP1OptV2(nq=961, nme=1800) run ( 1/ 1) : cputime=0.010683(s) - matrix 1922-by-1922 ... SquareMesh(30): -> StiffElasAssembling2DP1OptV2(nq=961, nme=1800) run ( 1/ 1) : cputime=0.010683(s) - matrix 1922-by-1922 ...
We also obtain
Benchmark of StiffElasAssembling2DP1 with versions OptV2 and OptV1
StiffElasAssembling2DP1 benchmark resume N nq ndof OptV2 OptV1 100 10201 20402 0.1110(s) 3.1286(s) x1.00 x28.18 200 40401 80802 0.5036(s) 12.5861(s) x1.00 x24.99 300 90601 181202 1.2058(s) 28.2232(s) x1.00 x23.41 400 160801 321602 2.1346(s) 50.2407(s) x1.00 x23.54 500 251001 502002 3.5360(s) 78.4291(s) x1.00 x22.18 600 361201 722402 5.3290(s) 113.4194(s) x1.00 x21.28 700 491401 982802 7.4691(s) 153.6793(s) x1.00 x20.58 800 641601 1283202 9.8943(s) 201.4319(s) x1.00 x20.36 This tabular was build with following code :
>>> from pyOptFEM.FEM2D import assemblyBench >>> assemblyBench(assembly='StiffElasAssembling2DP1',versions=['OptV2','OptV1'],LN=range(100,900,100)) SquareMesh(100): -> StiffElasAssembling2DP1OptV2(nq=10201, nme=20000) run ( 1/ 1) : cputime=0.111037(s) - matrix 20402-by-20402 ... SquareMesh(800): -> StiffElasAssembling2DP1OptV1(nq=641601, nme=1280000) run ( 1/ 1) : cputime=201.431866(s) - matrix 1283202-by-1283202 ...
We also obtain